1
2
3
4
5
6
7





Polycomplexes
C. elegans nuclei (blue) from animals lacking the essential axis component HTP-3. In these animals, SC material (magenta) forms chromosome-free aggregates called 'polycomplexes'. The essential crossover factor ZHP-3 (green) initially localizes to polycomplexes before concentrating in a small focus abutting polycomplexes.
Synapsed chromosomes
C. elegans gonad stained for SC and axis components (red and green, respectively). Colocalization of the green and red signals (resulting in yellow filaments) indicates synapsed homologous chromosomes. On the right side of the image some chromosomes begin to de-synapse, resulting in thinner green filaments.
Pairing Centers
C. elegans chromosomes have a specialized region - Pairing Center - that drives homologue pairing in early meiosis. A time-lapse series (top) shows that movements of the X chromosome (magenta) are mostly led by the pairing centers (green). Mapping the trajectories of multiple pairing centers on a representative nucleus (bottom) illustrates how their motions cover the entire nuclear envelope to help chromosomes find their homologous partner. (From Wynne et al., 2012)
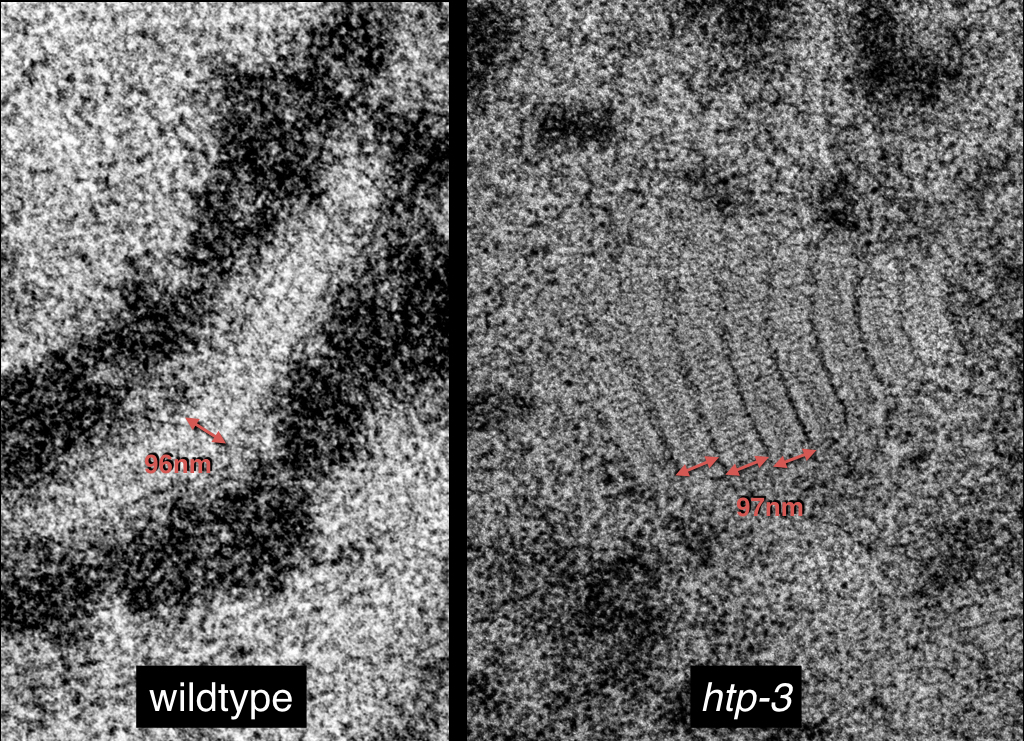
Polycomplexes
Electron microscopy reveals the ultrastructure of the synaptonemal complex and of polycomplexes. Left, synapsed chromosomes appear as elongated electron-dense structures that are connected through a 100nm-thick ladder-like synaptonemal complex. Right, when synaptonemal complex material cannot load onto chromosomes, it forms polycomplexes: ordered aggregates that resemble in their order and dimensions stacked synaptonemal complexes.
Synaptonemal Complex
C. elegans gonad stained for SC and axis components (red and green, respectively). Super-resolution imaging allows detection of two parallel axes, which are about ~100nm apart.